PDA22 - Proteomics Data Analysis 2022
DESCRIPTION
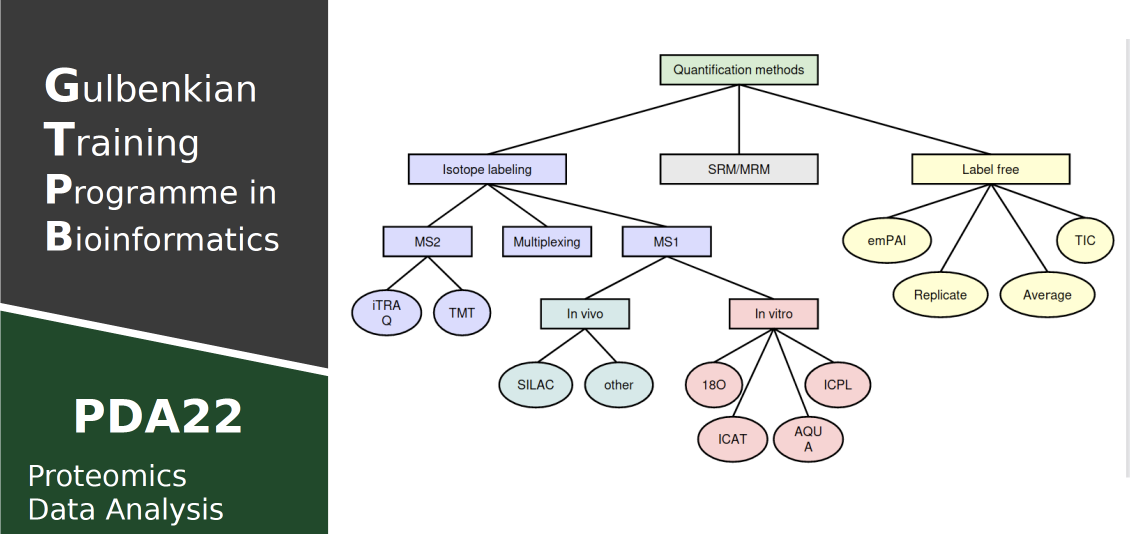
Mass spectrometry based proteomic experiments generate ever larger datasets and, as a consequence, complex data interpretation challenges. In this course, the concepts and methods required to tackle these challenges will be introduced, covering peptide and protein identification, quantification, and differential analysis. Moreover, more advanced experimental designs and blocking will also be introduced. The core focus will be on shotgun proteomics data, and quantification using label-free precursor peptide (MS1) ion intensities.
The course will rely exclusively on free and user-friendly software, all of which can be directly applied in your lab upon returning from the course. You will also learn how to submit data to PRIDE/ProteomeXchange, which is a common requirement for publication in the field, and how to browse and reprocess publicly available data from online repositories. The course will thus provide a solid basis for beginners, but will also bring new perspectives to those already familiar with standard d ata interpretation procedures in proteomics.
Note: This is a highly interactive course. It requires that the participants interact with each other and with the course instructors, in order to reach the learning outcomes in full.
INSTRUCTORS
Lennart Martens, Ghent University and VIB, Ghent, BE
Lieven Clement, Department of Applied Mathematics, Computer Science and Statistics, Ghent University, Ghent, BE
TARGET AUDIENCE
-
Ph.D. Students.
-
Biologists and bioinformaticians who are dealing with high-throughput gene expression data or other high-throughput data and would like to learn state-of-the-art methods for mining and analysing such data.
-
post-doctoral researchers and principle investigators working in wet-lab biology. Participants who are looking for a basic introduction to the bioinformatics resources offered by the EMBL-EBI and want to know more about accessing biological data and tools.
-
computational and statistical techniques are used to identify and validate the causal links between targets.
COURSE PRE-REQUISITES
The participants should have a basic knowledge about mass spectrometry based proteomics. Experience in analysing proteomics data is an advantage, but not mandatory. The course does not require advanced computer skills.
CAPACITY
20 participants
IMPORTANT DATES
Deadline for applications: September 5th, 2022
Course date: September 12th - September 16th, 2022
APPLICATION
More details about this course, including application instructions, are available at http://gtpb.igc.gulbenkian.pt/bicourses/2022/PDA22/
IGC - Instituto Gulbenkian de Ciência
Rua da Quinta Grande, 6
2780-156 Oeiras
Portugal